3.4 — Multivariate OLS Estimators
ECON 480 • Econometrics • Fall 2020
Ryan Safner
Assistant Professor of Economics
safner@hood.edu
ryansafner/metricsF20
metricsF20.classes.ryansafner.com
The Multivariate OLS Estimators
The Multivariate OLS Estimators
- By analogy, we still focus on the ordinary least squares (OLS) estimators of the unknown population parameters β0,β1,β2,⋯,βk which solves:
min^β0,^β1,^β2,⋯,^βkn∑i=1[Yi−(^β0+^β1X1i+^β2X2i+⋯+^βkXki)⏟ui]2
- Again, OLS estimators are chosen to minimize the sum of squared errors (SSE)
- i.e. sum of squared distances between actual values of Yi and predicted values ^Yi
The Multivariate OLS Estimators: FYI
Math FYI: in linear algebra terms, a regression model with n observations of k independent variables:
Y=Xβ+u
(y1y2⋮yn)⏟Y(n×1)=(x1,1x1,2⋯x1,nx2,1x2,2⋯x2,n⋮⋮⋱⋮xk,1xk,2⋯xk,n)⏟X(n×k)(β1β2⋮βk)⏟β(k×1)+(u1u2⋮un)⏟u(n×1)
The Multivariate OLS Estimators: FYI
Math FYI: in linear algebra terms, a regression model with n observations of k independent variables:
Y=Xβ+u
(y1y2⋮yn)⏟Y(n×1)=(x1,1x1,2⋯x1,nx2,1x2,2⋯x2,n⋮⋮⋱⋮xk,1xk,2⋯xk,n)⏟X(n×k)(β1β2⋮βk)⏟β(k×1)+(u1u2⋮un)⏟u(n×1)
- The OLS estimator for β is ˆβ=(X′X)−1X′Y 😱
The Multivariate OLS Estimators: FYI
Math FYI: in linear algebra terms, a regression model with n observations of k independent variables:
Y=Xβ+u
(y1y2⋮yn)⏟Y(n×1)=(x1,1x1,2⋯x1,nx2,1x2,2⋯x2,n⋮⋮⋱⋮xk,1xk,2⋯xk,n)⏟X(n×k)(β1β2⋮βk)⏟β(k×1)+(u1u2⋮un)⏟u(n×1)
- The OLS estimator for β is ˆβ=(X′X)−1X′Y 😱
- Appreciate that I am saving you from such sorrow 🤖
The Sampling Distribution of ^βj
- For any individual βj, it has a sampling distribution:
^βj∼N(E[^βj],se(^βj))
- We want to know its sampling distribution’s:
- Center: E[^βj]; what is the expected value of our estimator?
- Spread: se(^βj); how precise or uncertain is our estimator?
The Sampling Distribution of ^βj
- For any individual βj, it has a sampling distribution:
^βj∼N(E[^βj],se(^βj))
- We want to know its sampling distribution’s:
- Center: E[^βj]; what is the expected value of our estimator?
- Spread: se(^βj); how precise or uncertain is our estimator?

The Expected Value of ^βj: Bias
Exogeneity and Unbiasedness
- As before, E[^βj]=βj when Xj is exogenous (i.e. cor(Xj,u)=0)
Exogeneity and Unbiasedness
As before, E[^βj]=βj when Xj is exogenous (i.e. cor(Xj,u)=0)
We know the true E[^βj]=βj+cor(Xj,u)σuσXj⏟O.V. Bias
Exogeneity and Unbiasedness
As before, E[^βj]=βj when Xj is exogenous (i.e. cor(Xj,u)=0)
We know the true E[^βj]=βj+cor(Xj,u)σuσXj⏟O.V. Bias
If Xj is endogenous (i.e. cor(Xj,u)≠0), contains omitted variable bias
Exogeneity and Unbiasedness
As before, E[^βj]=βj when Xj is exogenous (i.e. cor(Xj,u)=0)
We know the true E[^βj]=βj+cor(Xj,u)σuσXj⏟O.V. Bias
If Xj is endogenous (i.e. cor(Xj,u)≠0), contains omitted variable bias
We can now try to quantify the omitted variable bias
Measuring Omitted Variable Bias I
- Suppose the true population model of a relationship is:
Yi=β0+β1X1i+β2X2i+ui
- What happens when we run a regression and omit X2i?
Measuring Omitted Variable Bias I
- Suppose the true population model of a relationship is:
Yi=β0+β1X1i+β2X2i+ui
What happens when we run a regression and omit X2i?
Suppose we estimate the following omitted regression of just Yi on X1i (omitting X2i):†
Yi=α0+α1X1i+νi
† Note: I am using α's and νi only to denote these are different estimates than the true model β's and ui
Measuring Omitted Variable Bias II
- Key Question: are X1i and X2i correlated?
Measuring Omitted Variable Bias II
Key Question: are X1i and X2i correlated?
Run an auxiliary regression of X2i on X1i to see:†
X2i=δ0+δ1X1i+τi
Measuring Omitted Variable Bias II
Key Question: are X1i and X2i correlated?
Run an auxiliary regression of X2i on X1i to see:†
X2i=δ0+δ1X1i+τi
If δ1=0, then X1i and X2i are not linearly related
If |δ1| is very big, then X1i and X2i are strongly linearly related
† Note: I am using δ's and τ to differentiate estimates for this model.
Measuring Omitted Variable Bias III
- Now substitute our auxiliary regression between X2i and X1i into the true model:
- We know X2i=δ0+δ1X1i+τi
Yi=β0+β1X1i+β2X2i+ui
Measuring Omitted Variable Bias III
- Now substitute our auxiliary regression between X2i and X1i into the true model:
- We know X2i=δ0+δ1X1i+τi
Yi=β0+β1X1i+β2X2i+uiYi=β0+β1X1i+β2(δ0+δ1X1i+τi)+ui
Measuring Omitted Variable Bias III
- Now substitute our auxiliary regression between X2i and X1i into the true model:
- We know X2i=δ0+δ1X1i+τi
Yi=β0+β1X1i+β2X2i+uiYi=β0+β1X1i+β2(δ0+δ1X1i+τi)+uiYi=(β0+β2δ0)+(β1+β2δ1)X1i+(β2τi+ui)
Measuring Omitted Variable Bias III
- Now substitute our auxiliary regression between X2i and X1i into the true model:
- We know X2i=δ0+δ1X1i+τi
Yi=β0+β1X1i+β2X2i+uiYi=β0+β1X1i+β2(δ0+δ1X1i+τi)+uiYi=(β0+β2δ0⏟α0)+(β1+β2δ1⏟α1)X1i+(β2τi+ui⏟νi)
- Now relabel each of the three terms as the OLS estimates (α's) and error (νi) from the omitted regression, so we again have:
Yi=α0+α1X1i+νi
Measuring Omitted Variable Bias III
- Now substitute our auxiliary regression between X2i and X1i into the true model:
- We know X2i=δ0+δ1X1i+τi
Yi=β0+β1X1i+β2X2i+uiYi=β0+β1X1i+β2(δ0+δ1X1i+τi)+uiYi=(β0+β2δ0⏟α0)+(β1+β2δ1⏟α1)X1i+(β2τi+ui⏟νi)
- Now relabel each of the three terms as the OLS estimates (α's) and error (νi) from the omitted regression, so we again have:
Yi=α0+α1X1i+νi
- Crucially, this means that our OLS estimate for X1i in the omitted regression is:
α1=β1+β2δ1
Measuring Omitted Variable Bias IV
α1=β1 + β2δ1
- The Omitted Regression OLS estimate for X1i, (α1) picks up both:
Measuring Omitted Variable Bias IV
α1=β1 + β2δ1
- The Omitted Regression OLS estimate for X1i, (α1) picks up both:
- The true effect of X1 on Yi: (β1)
Measuring Omitted Variable Bias IV
α1=β1 + β2δ1
- The Omitted Regression OLS estimate for X1i, (α1) picks up both:
- The true effect of X1 on Yi: (β1)
- The true effect of X2 on Yi: (β2)
- As pulled through the relationship between X1 and X2: (δ1)
Measuring Omitted Variable Bias IV
α1=β1 + β2δ1
- The Omitted Regression OLS estimate for X1i, (α1) picks up both:
- The true effect of X1 on Yi: (β1)
- The true effect of X2 on Yi: (β2)
- As pulled through the relationship between X1 and X2: (δ1)
- Recall our conditions for omitted variable bias from some variable Zi:
Measuring Omitted Variable Bias IV
α1=β1 + β2δ1
- The Omitted Regression OLS estimate for X1i, (α1) picks up both:
- The true effect of X1 on Yi: (β1)
- The true effect of X2 on Yi: (β2)
- As pulled through the relationship between X1 and X2: (δ1)
- Recall our conditions for omitted variable bias from some variable Zi:
1) Zi must be a determinant of Yi ⟹ β2≠0
Measuring Omitted Variable Bias IV
α1=β1 + β2δ1
- The Omitted Regression OLS estimate for X1i, (α1) picks up both:
- The true effect of X1 on Yi: (β1)
- The true effect of X2 on Yi: (β2)
- As pulled through the relationship between X1 and X2: (δ1)
- Recall our conditions for omitted variable bias from some variable Zi:
1) Zi must be a determinant of Yi ⟹ β2≠0
2) Zi must be correlated with Xi ⟹ δ1≠0
Measuring Omitted Variable Bias IV
α1=β1 + β2δ1
- The Omitted Regression OLS estimate for X1i, (α1) picks up both:
- The true effect of X1 on Yi: (β1)
- The true effect of X2 on Yi: (β2)
- As pulled through the relationship between X1 and X2: (δ1)
- Recall our conditions for omitted variable bias from some variable Zi:
1) Zi must be a determinant of Yi ⟹ β2≠0
2) Zi must be correlated with Xi ⟹ δ1≠0
- Otherwise, if Zi does not fit these conditions, α1=β1 and the omitted regression is unbiased!
Measuring OVB in Our Class Size Example I
- The “True” Regression (Yi on X1i and X2i)
^Test Scorei=686.03−1.10 STRi−0.65 %ELi
| ABCDEFGHIJ0123456789 |
term <chr> | estimate <dbl> | std.error <dbl> | statistic <dbl> | p.value <dbl> |
|---|---|---|---|---|
| (Intercept) | 686.0322487 | 7.41131248 | 92.565554 | 3.871501e-280 |
| str | -1.1012959 | 0.38027832 | -2.896026 | 3.978056e-03 |
| el_pct | -0.6497768 | 0.03934255 | -16.515879 | 1.657506e-47 |
Measuring OVB in Our Class Size Example II
- The “Omitted” Regression (Yi on just X1i)
^Test Scorei=698.93−2.28 STRi
| ABCDEFGHIJ0123456789 |
term <chr> | estimate <dbl> | std.error <dbl> | statistic <dbl> | p.value <dbl> |
|---|---|---|---|---|
| (Intercept) | 698.932952 | 9.4674914 | 73.824514 | 6.569925e-242 |
| str | -2.279808 | 0.4798256 | -4.751327 | 2.783307e-06 |
Measuring OVB in Our Class Size Example III
- The “Auxiliary” Regression (X2i on X1i)
^%ELi=−19.85+1.81 STRi
| ABCDEFGHIJ0123456789 |
term <chr> | estimate <dbl> | std.error <dbl> | statistic <dbl> | p.value <dbl> |
|---|---|---|---|---|
| (Intercept) | -19.854055 | 9.1626044 | -2.166857 | 0.0308099863 |
| str | 1.813719 | 0.4643735 | 3.905733 | 0.0001095165 |
Measuring OVB in Our Class Size Example IV
“True” Regression
^Test Scorei=686.03−1.10 STRi−0.65 %EL
“Omitted” Regression
^Test Scorei=698.93−2.28 STRi
“Auxiliary” Regression
^%ELi=−19.85+1.81 STRi
- Omitted Regression α1 on STR is -2.28
Measuring OVB in Our Class Size Example IV
“True” Regression
^Test Scorei=686.03−1.10 STRi−0.65 %EL
“Omitted” Regression
^Test Scorei=698.93−2.28 STRi
“Auxiliary” Regression
^%ELi=−19.85+1.81 STRi
- Omitted Regression α1 on STR is -2.28
α1=β1+β2δ1
- The true effect of STR on Test Score: -1.10
Measuring OVB in Our Class Size Example IV
“True” Regression
^Test Scorei=686.03−1.10 STRi−0.65 %EL
“Omitted” Regression
^Test Scorei=698.93−2.28 STRi
“Auxiliary” Regression
^%ELi=−19.85+1.81 STRi
- Omitted Regression α1 on STR is -2.28
α1=β1+β2δ1
The true effect of STR on Test Score: -1.10
The true effect of %EL on Test Score: -0.65
Measuring OVB in Our Class Size Example IV
“True” Regression
^Test Scorei=686.03−1.10 STRi−0.65 %EL
“Omitted” Regression
^Test Scorei=698.93−2.28 STRi
“Auxiliary” Regression
^%ELi=−19.85+1.81 STRi
- Omitted Regression α1 on STR is -2.28
α1=β1+β2δ1
The true effect of STR on Test Score: -1.10
The true effect of %EL on Test Score: -0.65
The relationship between STR and %EL: 1.81
Measuring OVB in Our Class Size Example IV
“True” Regression
^Test Scorei=686.03−1.10 STRi−0.65 %EL
“Omitted” Regression
^Test Scorei=698.93−2.28 STRi
“Auxiliary” Regression
^%ELi=−19.85+1.81 STRi
- Omitted Regression α1 on STR is -2.28
α1=β1+β2δ1
The true effect of STR on Test Score: -1.10
The true effect of %EL on Test Score: -0.65
The relationship between STR and %EL: 1.81
So, for the omitted regression:
−2.28=−1.10+(−0.65)(1.81)
Measuring OVB in Our Class Size Example IV
“True” Regression
^Test Scorei=686.03−1.10 STRi−0.65 %EL
“Omitted” Regression
^Test Scorei=698.93−2.28 STRi
“Auxiliary” Regression
^%ELi=−19.85+1.81 STRi
- Omitted Regression α1 on STR is -2.28
α1=β1+β2δ1
The true effect of STR on Test Score: -1.10
The true effect of %EL on Test Score: -0.65
The relationship between STR and %EL: 1.81
So, for the omitted regression:
−2.28=−1.10+(−0.65)(1.81)⏟O.V.Bias=−1.18
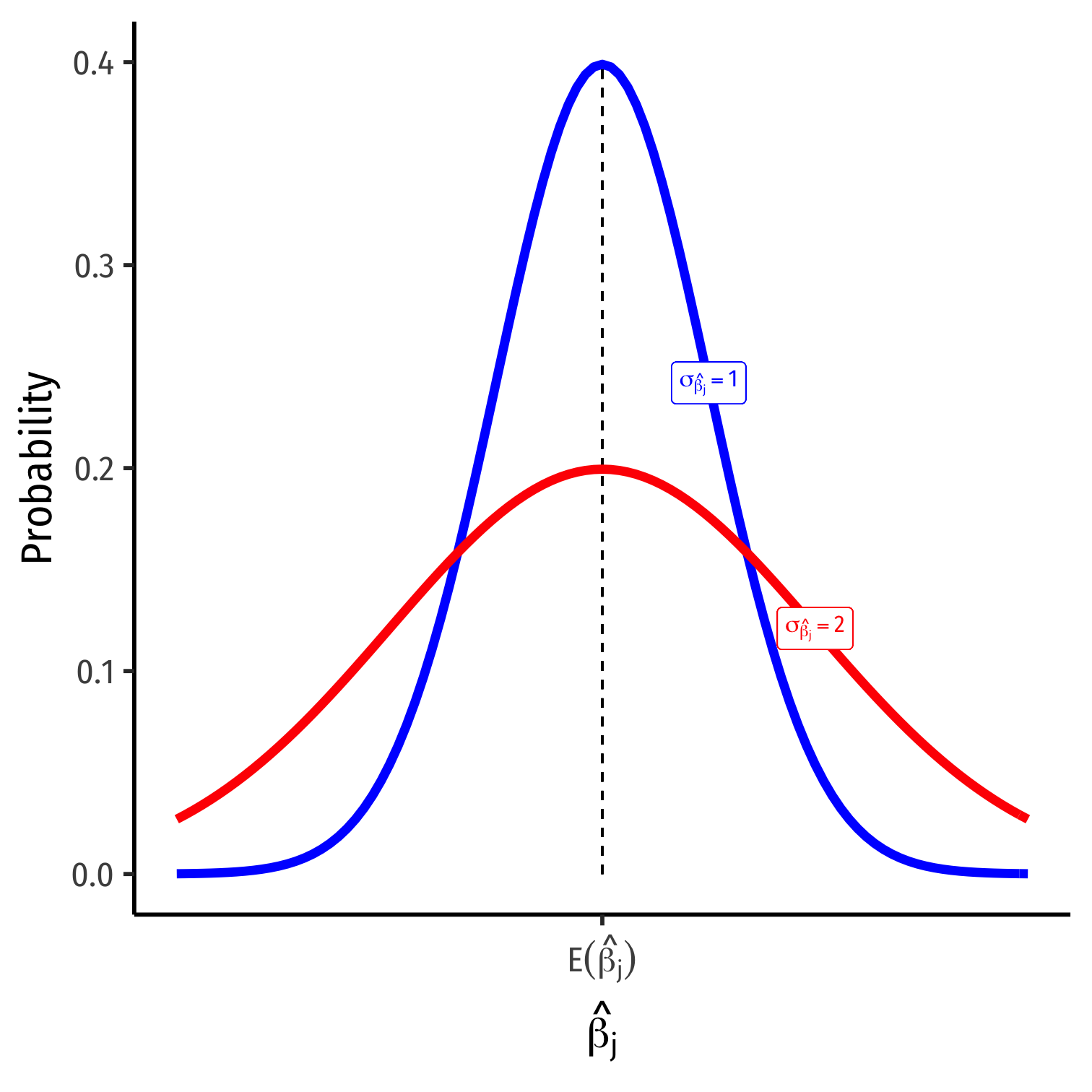
Precision of ^βj
Precision of ^βj I
σ^βj; how precise are our estimates?
Variance σ2^βj or standard error σ^βj
Precision of ^βj II
var(^βj)=11−R2j⏟VIF×(SER)2n×var(X)
se(^βj)=√var(^β1)
- Variation in ^βj is affected by four things now†:
- Goodness of fit of the model (SER)
- Larger SER → larger var(^βj)
- Sample size, n
- Larger n → smaller var(^βj)
- Variance of X
- Larger var(X) → smaller var(^βj)
- Variance Inflation Factor 1(1−R2j)
- Larger VIF, larger var(^βj)
- This is the only new effect
† See Class 2.5 for a reminder of variation with just one X variable.
VIF and Multicollinearity I
- Two independent variables are multicollinear:
cor(Xj,Xl)≠0∀j≠l
VIF and Multicollinearity I
- Two independent variables are multicollinear:
cor(Xj,Xl)≠0∀j≠l
- Multicollinearity between X variables does not bias OLS estimates
- Remember, we pulled another variable out of u into the regression
- If it were omitted, then it would cause omitted variable bias!
VIF and Multicollinearity I
- Two independent variables are multicollinear:
cor(Xj,Xl)≠0∀j≠l
Multicollinearity between X variables does not bias OLS estimates
- Remember, we pulled another variable out of u into the regression
- If it were omitted, then it would cause omitted variable bias!
Multicollinearity does increase the variance of each estimate by
VIF=1(1−R2j)
VIF and Multicollinearity II
VIF=1(1−R2j)
- R2j is the R2 from an auxiliary regression of Xj on all other regressors (X’s)
VIF and Multicollinearity II
VIF=1(1−R2j)
- R2j is the R2 from an auxiliary regression of Xj on all other regressors (X’s)
Example: Suppose we have a regression with three regressors (k=3):
Yi=β0+β1X1i+β2X2i+β3X3i
VIF and Multicollinearity II
VIF=1(1−R2j)
- R2j is the R2 from an auxiliary regression of Xj on all other regressors (X’s)
Example: Suppose we have a regression with three regressors (k=3):
Yi=β0+β1X1i+β2X2i+β3X3i
- There will be three different R2j's, one for each regressor:
R21 for X1i=γ+γX2i+γX3iR22 for X2i=ζ0+ζ1X1i+ζ2X3iR23 for X3i=η0+η1X1i+η2X2i
VIF and Multicollinearity III
VIF=1(1−R2j)
R2j is the R2 from an auxiliary regression of Xj on all other regressors (X's)
The R2j tells us how much other regressors explain regressor Xj
Key Takeaway: If other X variables explain Xj well (high R2J), it will be harder to tell how cleanly Xj→Yi, and so var(^βj) will be higher
VIF and Multicollinearity IV
- Common to calculate the Variance Inflation Factor (VIF) for each regressor:
VIF=1(1−R2j)
- VIF quantifies the factor (scalar) by which var(^βj) increases because of multicollinearity
- e.g. VIF of 2, 3, etc. ⟹ variance increases by 2x, 3x, etc.
VIF and Multicollinearity IV
- Common to calculate the Variance Inflation Factor (VIF) for each regressor:
VIF=1(1−R2j)
- VIF quantifies the factor (scalar) by which var(^βj) increases because of multicollinearity
- e.g. VIF of 2, 3, etc. ⟹ variance increases by 2x, 3x, etc.
- Baseline: R2j=0 ⟹ no multicollinearity ⟹VIF=1 (no inflation)
VIF and Multicollinearity IV
- Common to calculate the Variance Inflation Factor (VIF) for each regressor:
VIF=1(1−R2j)
- VIF quantifies the factor (scalar) by which var(^βj) increases because of multicollinearity
- e.g. VIF of 2, 3, etc. ⟹ variance increases by 2x, 3x, etc.
Baseline: R2j=0 ⟹ no multicollinearity ⟹VIF=1 (no inflation)
Larger R2j ⟹ larger VIF
- Rule of thumb: VIF>10 is problematic
VIF and Multicollinearity V
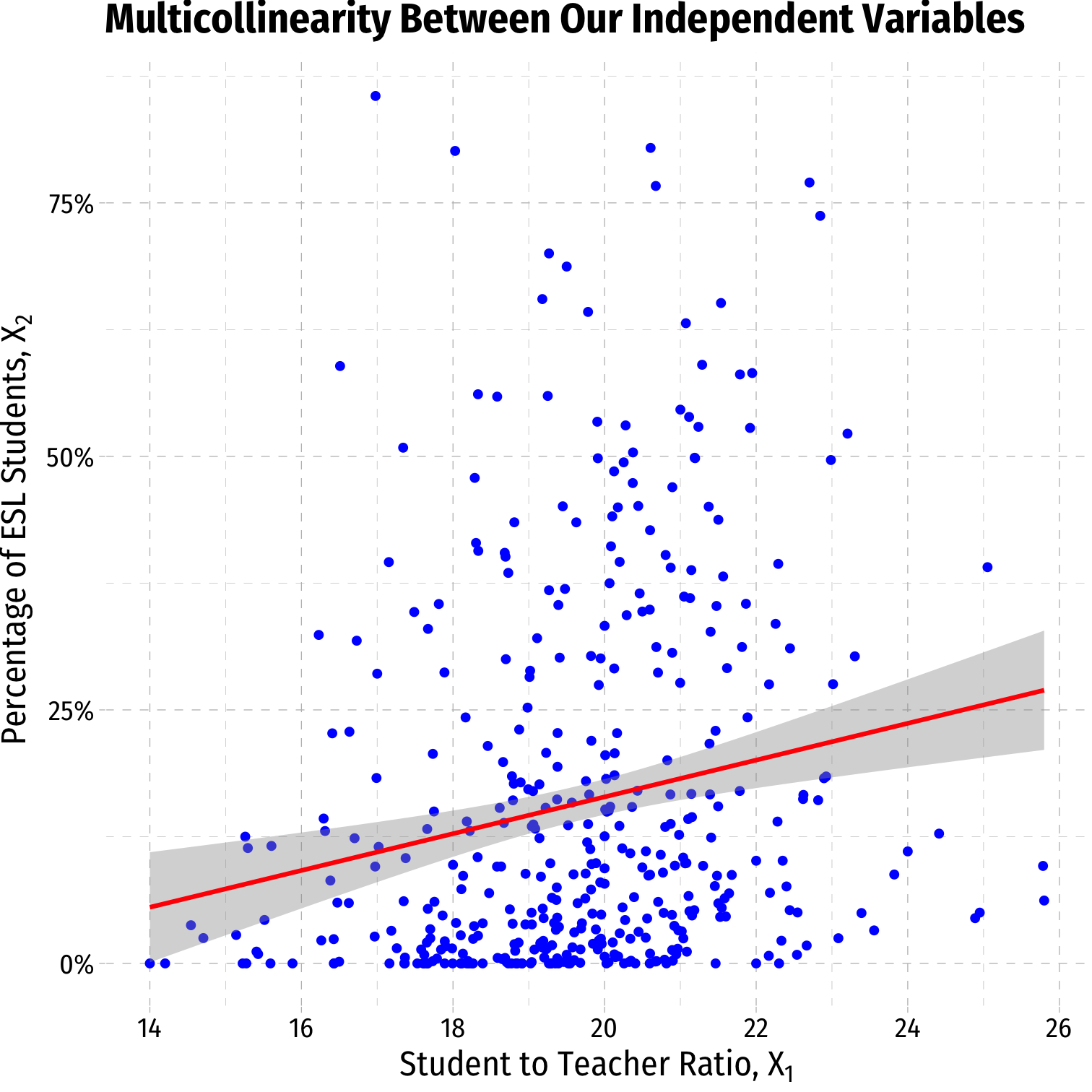
# scatterplot of X2 on X1ggplot(data=CASchool, aes(x=str,y=el_pct))+ geom_point(color="blue")+ geom_smooth(method="lm", color="red")+ scale_y_continuous(labels=function(x){paste0(x,"%")})+ labs(x = expression(paste("Student to Teacher Ratio, ", X[1])), y = expression(paste("Percentage of ESL Students, ", X[2])), title = "Multicollinearity Between Our Independent Variables")+ ggthemes::theme_pander(base_family = "Fira Sans Condensed", base_size=16)# Make a correlation tableCASchool %>% select(testscr, str, el_pct) %>% cor()## testscr str el_pct## testscr 1.0000000 -0.2263628 -0.6441237## str -0.2263628 1.0000000 0.1876424## el_pct -0.6441237 0.1876424 1.0000000- Cor(STR, %EL) = -0.644
VIF and Multicollinearity in R I
# our multivariate regressionelreg <- lm(testscr ~ str + el_pct, data = CASchool)# use the "car" package for VIF function library("car") # syntax: vif(lm.object)vif(elreg)## str el_pct ## 1.036495 1.036495VIF and Multicollinearity in R I
# our multivariate regressionelreg <- lm(testscr ~ str + el_pct, data = CASchool)# use the "car" package for VIF function library("car") # syntax: vif(lm.object)vif(elreg)## str el_pct ## 1.036495 1.036495- var(^β1) on
strincreases by 1.036 times due to multicollinearity withel_pct - var(^β2) on
el_pctincreases by 1.036 times due to multicollinearity withstr
VIF and Multicollinearity in R II
- Let's calculate VIF manually to see where it comes from:
VIF and Multicollinearity in R II
- Let's calculate VIF manually to see where it comes from:
# run auxiliary regression of x2 on x1auxreg <- lm(el_pct ~ str, data = CASchool)# use broom package's tidy() command (cleaner)library(broom) # load broomtidy(auxreg) # look at reg output| ABCDEFGHIJ0123456789 |
term <chr> | estimate <dbl> | std.error <dbl> | statistic <dbl> | p.value <dbl> |
|---|---|---|---|---|
| (Intercept) | -19.854055 | 9.1626044 | -2.166857 | 0.0308099863 |
| str | 1.813719 | 0.4643735 | 3.905733 | 0.0001095165 |
VIF and Multicollinearity in R III
glance(auxreg) # look at aux reg stats for R^2| ABCDEFGHIJ0123456789 |
r.squared <dbl> | adj.r.squared <dbl> | sigma <dbl> | statistic <dbl> | p.value <dbl> | df <dbl> | |
|---|---|---|---|---|---|---|
| 0.03520966 | 0.03290155 | 17.98259 | 15.25475 | 0.0001095165 | 1 |
# extract our R-squared from aux regression (R_j^2)aux_r_sq<-glance(auxreg) %>%
select(r.squared)aux_r_sq # look at it
| ABCDEFGHIJ0123456789 |
r.squared <dbl> | ||||
|---|---|---|---|---|
| 0.03520966 |
VIF and Multicollinearity in R IV
# calculate VIF manuallyour_vif<-1/(1-aux_r_sq) # VIF formula our_vif| ABCDEFGHIJ0123456789 |
r.squared <dbl> | ||||
|---|---|---|---|---|
| 1.036495 |
- Again, multicollinearity between the two X variables inflates the variance on each by 1.036 times
VIF and Multicollinearity: Another Example I
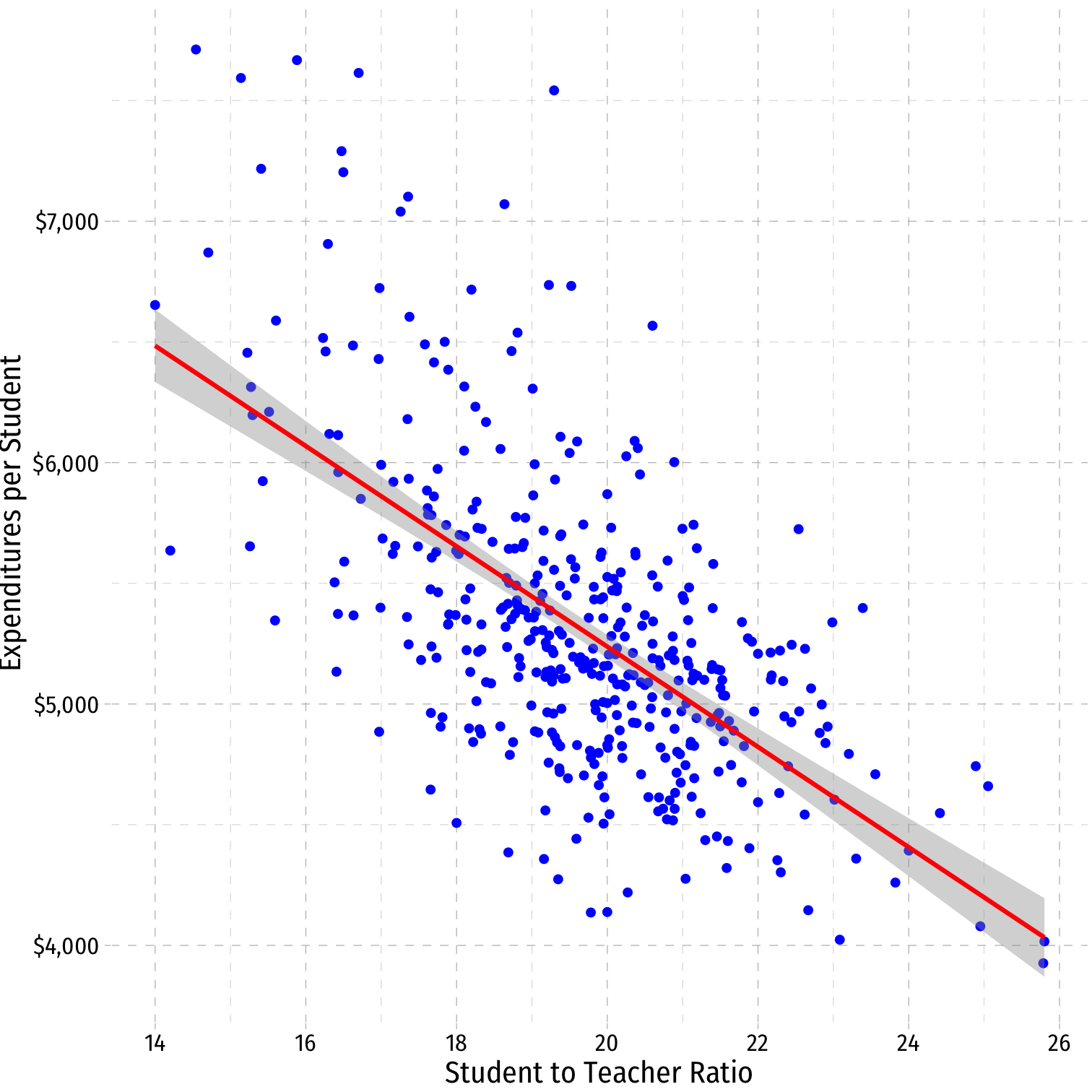
Example: What about district expenditures per student?
CASchool %>% select(testscr, str, expn_stu) %>% cor()## testscr str expn_stu## testscr 1.0000000 -0.2263628 0.1912728## str -0.2263628 1.0000000 -0.6199821## expn_stu 0.1912728 -0.6199821 1.0000000VIF and Multicollinearity: Another Example II
ggplot(data=CASchool, aes(x=str,y=expn_stu))+ geom_point(color="blue")+ geom_smooth(method="lm", color="red")+ scale_y_continuous(labels = scales::dollar)+ labs(x = "Student to Teacher Ratio", y = "Expenditures per Student")+ ggthemes::theme_pander(base_family = "Fira Sans Condensed", base_size=14)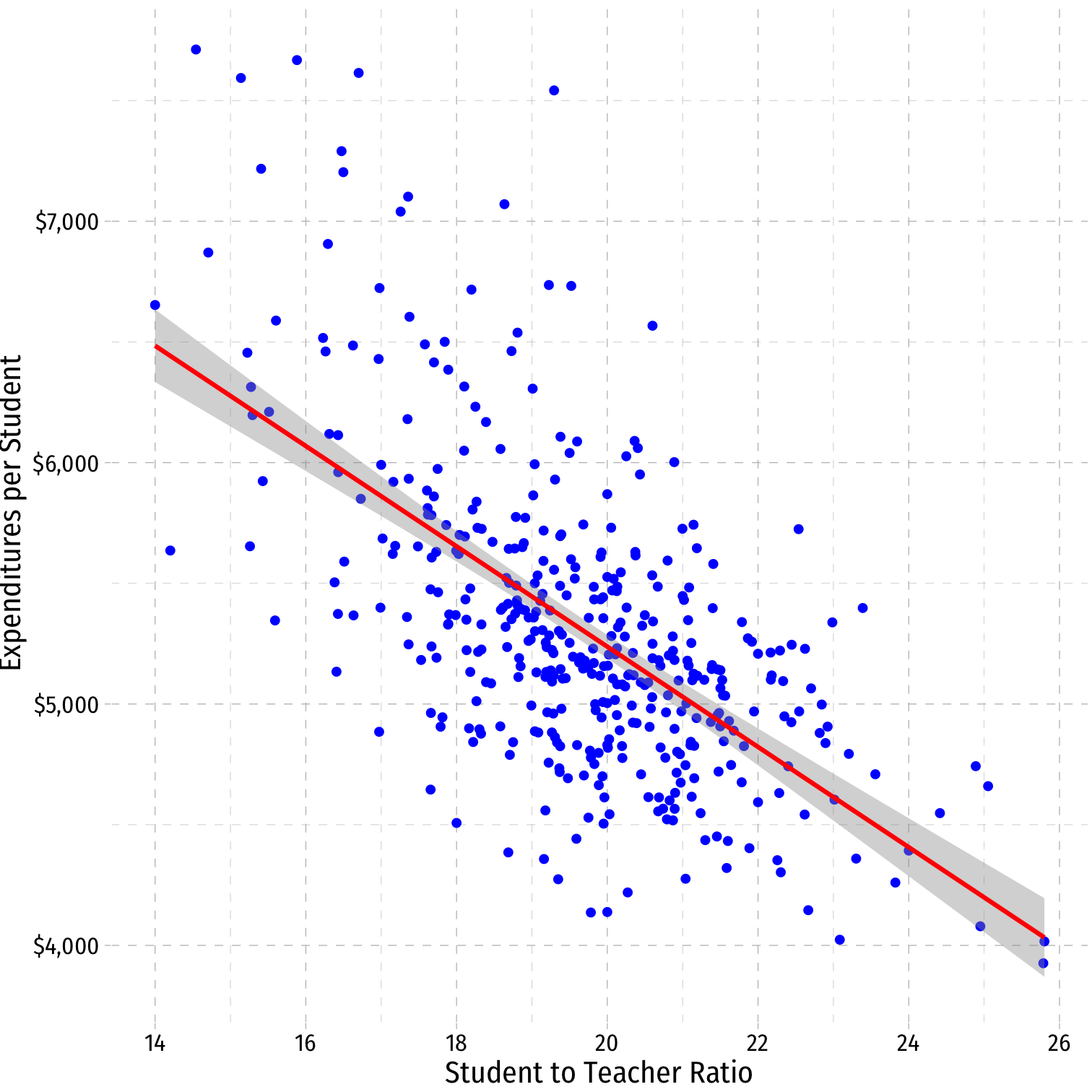
VIF and Multicollinearity: Another Example III
cor(Test score, expn)≠0
cor(STR, expn)≠0
VIF and Multicollinearity: Another Example III
cor(Test score, expn)≠0
cor(STR, expn)≠0
- Omitting expn will bias ^β1 on STR
VIF and Multicollinearity: Another Example III
cor(Test score, expn)≠0
cor(STR, expn)≠0
Omitting expn will bias ^β1 on STR
Including expn will not bias ^β1 on STR, but will make it less precise (higher variance)
VIF and Multicollinearity: Another Example III
- Data tells us little about the effect of a change in STR holding expn constant
- Hard to know what happens to test scores when high STR AND high expn and vice versa (they rarely happen simultaneously)!
VIF and Multicollinearity: Another Example IV
expreg <- lm(testscr ~ str + expn_stu, data = CASchool)expreg %>% tidy()| ABCDEFGHIJ0123456789 |
term <chr> | estimate <dbl> | |
|---|---|---|
| (Intercept) | 675.577173851 | |
| str | -1.763215599 | |
| expn_stu | 0.002486571 |
VIF and Multicollinearity: Another Example IV
expreg <- lm(testscr ~ str + expn_stu, data = CASchool)expreg %>% tidy()| ABCDEFGHIJ0123456789 |
term <chr> | estimate <dbl> | |
|---|---|---|
| (Intercept) | 675.577173851 | |
| str | -1.763215599 | |
| expn_stu | 0.002486571 |
expreg %>% vif()## str expn_stu ## 1.624373 1.624373- Including
expn_stuincreases variance of ^β1 and ^β2 by 1.62x (62%)
Multicollinearity Increases Variance
library(huxtable)huxreg("Model 1" = school_reg, "Model 2" = expreg, coefs = c("Intercept" = "(Intercept)", "Class Size" = "str", "Expenditures per Student" = "expn_stu"), statistics = c("N" = "nobs", "R-Squared" = "r.squared", "SER" = "sigma"), number_format = 2)- We can see SE(^β1) on
strincreases from 0.48 to 0.61 when we addexpn_stu
| Model 1 | Model 2 | |
|---|---|---|
| Intercept | 698.93 *** | 675.58 *** |
| (9.47) | (19.56) | |
| Class Size | -2.28 *** | -1.76 ** |
| (0.48) | (0.61) | |
| Expenditures per Student | 0.00 | |
| (0.00) | ||
| N | 420 | 420 |
| R-Squared | 0.05 | 0.06 |
| SER | 18.58 | 18.56 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | ||
Perfect Multicollinearity
- Perfect multicollinearity is when a regressor is an exact linear function of (an)other regressor(s)
Perfect Multicollinearity
- Perfect multicollinearity is when a regressor is an exact linear function of (an)other regressor(s)
^Sales=^β0+^β1Temperature (C)+^β2Temperature (F)
Perfect Multicollinearity
- Perfect multicollinearity is when a regressor is an exact linear function of (an)other regressor(s)
^Sales=^β0+^β1Temperature (C)+^β2Temperature (F)
Temperature (F)=32+1.8∗Temperature (C)
Perfect Multicollinearity
- Perfect multicollinearity is when a regressor is an exact linear function of (an)other regressor(s)
^Sales=^β0+^β1Temperature (C)+^β2Temperature (F)
Temperature (F)=32+1.8∗Temperature (C)
- cor(temperature (F), temperature (C))=1
Perfect Multicollinearity
- Perfect multicollinearity is when a regressor is an exact linear function of (an)other regressor(s)
^Sales=^β0+^β1Temperature (C)+^β2Temperature (F)
Temperature (F)=32+1.8∗Temperature (C)
cor(temperature (F), temperature (C))=1
R2j=1 is implying VIF=11−1 and var(^βj)=0!
Perfect Multicollinearity
- Perfect multicollinearity is when a regressor is an exact linear function of (an)other regressor(s)
^Sales=^β0+^β1Temperature (C)+^β2Temperature (F)
Temperature (F)=32+1.8∗Temperature (C)
cor(temperature (F), temperature (C))=1
R2j=1 is implying VIF=11−1 and var(^βj)=0!
This is fatal for a regression
- A logical impossiblity, always caused by human error
Perfect Multicollinearity: Example
Example:
^TestScorei=^β0+^β1STRi+^β2%EL+^β3%EF
%EL: the percentage of students learning English
%EF: the percentage of students fluent in English
EF=100−EL
|cor(EF,EL)|=1
Perfect Multicollinearity Example II
# generate %EF variable from %ELCASchool_ex <- CASchool %>% mutate(ef_pct = 100 - el_pct)# get correlation between %EL and %EFCASchool_ex %>% summarize(cor = cor(ef_pct, el_pct))| cor |
|---|
| -1 |
Perfect Multicollinearity Example III
ggplot(data=CASchool_ex, aes(x=el_pct,y=ef_pct))+ geom_point(color="blue")+ scale_y_continuous(labels = scales::dollar)+ labs(x = "Percent of ESL Students", y = "Percent of Non-ESL Students")+ ggthemes::theme_pander(base_family = "Fira Sans Condensed", base_size=16)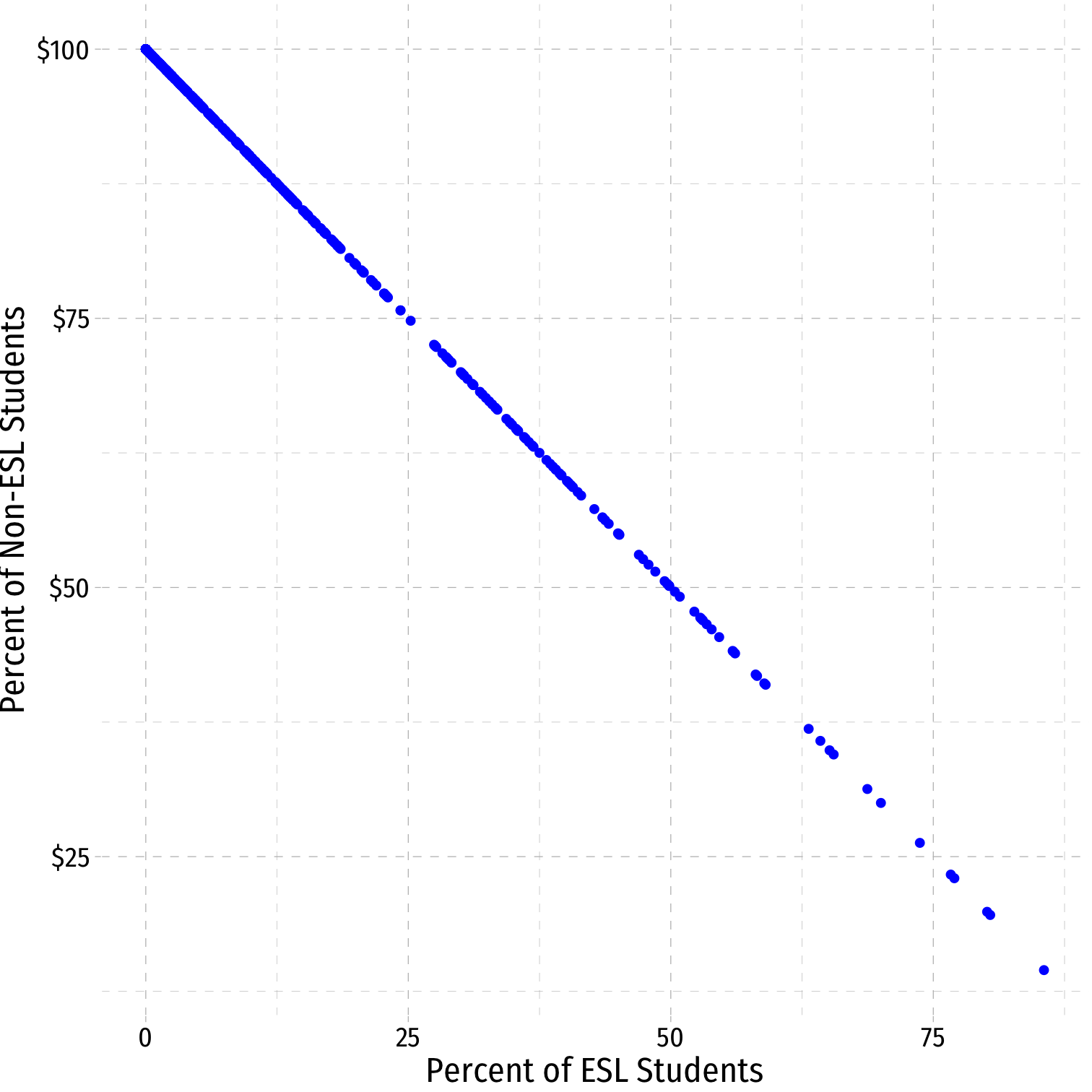
Perfect Multicollinearity Example IV
mcreg <- lm(testscr ~ str + el_pct + ef_pct, data = CASchool_ex)summary(mcreg)## ## Call:## lm(formula = testscr ~ str + el_pct + ef_pct, data = CASchool_ex)## ## Residuals:## Min 1Q Median 3Q Max ## -48.845 -10.240 -0.308 9.815 43.461 ## ## Coefficients: (1 not defined because of singularities)## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 686.03225 7.41131 92.566 < 2e-16 ***## str -1.10130 0.38028 -2.896 0.00398 ** ## el_pct -0.64978 0.03934 -16.516 < 2e-16 ***## ef_pct NA NA NA NA ## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1## ## Residual standard error: 14.46 on 417 degrees of freedom## Multiple R-squared: 0.4264, Adjusted R-squared: 0.4237 ## F-statistic: 155 on 2 and 417 DF, p-value: < 2.2e-16mcreg %>% tidy()| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | 686 | 7.41 | 92.6 | 3.87e-280 |
| str | -1.1 | 0.38 | -2.9 | 0.00398 |
| el_pct | -0.65 | 0.0393 | -16.5 | 1.66e-47 |
| ef_pct |
- Note
Rdrops one of the multicollinear regressors (ef_pct) if you include both 🤡
A Summary of Multivariate OLS Estimator Properties
A Summary of Multivariate OLS Estimator Properties
^βj on Xj is biased only if there is an omitted variable (Z) such that:
- cor(Y,Z)≠0
- cor(Xj,Z)≠0
- If Z is included and Xj is collinear with Z, this does not cause a bias
var[^βj] and se[^βj] measure precision (or uncertainty) of estimate:
A Summary of Multivariate OLS Estimator Properties
^βj on Xj is biased only if there is an omitted variable (Z) such that:
- cor(Y,Z)≠0
- cor(Xj,Z)≠0
- If Z is included and Xj is collinear with Z, this does not cause a bias
var[^βj] and se[^βj] measure precision (or uncertainty) of estimate:
var[^βj]=1(1−R2j)∗SER2n×var[Xj]
- VIF from multicollinearity: 1(1−R2j)
- R2j for auxiliary regression of Xj on all other X's
- mutlicollinearity does not bias ^βj but raises its variance
- perfect multicollinearity if X's are linear function of others
Updated Measures of Fit
(Updated) Measures of Fit
Again, how well does a linear model fit the data?
How much variation in Yi is “explained” by variation in the model (^Yi)?
(Updated) Measures of Fit
Again, how well does a linear model fit the data?
How much variation in Yi is “explained” by variation in the model (^Yi)?
Yi=^Yi+^ui^ui=Yi−^Yi
(Updated) Measures of Fit: SER
- Again, the Standard errror of the regression (SER) estimates the standard error of u
SER=SSEn−k−1
A measure of the spread of the observations around the regression line (in units of Y), the average "size" of the residual
Only new change: divided by n−k−1 due to use of k+1 degrees of freedom to first estimate β0 and then all of the other β's for the k number of regressors†
† Again, because your textbook defines k as including the constant, the denominator would be n-k instead of n-k-1.
(Updated) Measures of Fit: R2
R2=ESSTSS=1−SSETSS=(rX,Y)2
- Again, R2 is fraction of variation of the model (^Yi (“explained sum of squares”) to the variation of observations of Yi (“total sum of squares”)
(Updated) Measures of Fit: Adjusted ˉR2
Problem: R2 of a regression increases every time a new variable is added (it reduces SSE!)
This does not mean adding a variable improves the fit of the model per se, R2 gets inflated
(Updated) Measures of Fit: Adjusted ˉR2
Problem: R2 of a regression increases every time a new variable is added (it reduces SSE!)
This does not mean adding a variable improves the fit of the model per se, R2 gets inflated
We correct for this effect with the adjusted R2:
ˉR2=1−n−1n−k−1×SSETSS
- There are different methods to compute ˉR2, and in the end, recall R2 was never very useful, so don't worry about knowing the formula
- Large sample sizes (n) make R2 and ˉR2 very close
In R (base)
## ## Call:## lm(formula = testscr ~ str + el_pct, data = CASchool)## ## Residuals:## Min 1Q Median 3Q Max ## -48.845 -10.240 -0.308 9.815 43.461 ## ## Coefficients:## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 686.03225 7.41131 92.566 < 2e-16 ***## str -1.10130 0.38028 -2.896 0.00398 ** ## el_pct -0.64978 0.03934 -16.516 < 2e-16 ***## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1## ## Residual standard error: 14.46 on 417 degrees of freedom## Multiple R-squared: 0.4264, Adjusted R-squared: 0.4237 ## F-statistic: 155 on 2 and 417 DF, p-value: < 2.2e-16- Base R2 (
Rcalls it “Multiple R-squared”) went up Adjusted R-squaredwent down
In R (broom)
elreg %>% glance()| r.squared | adj.r.squared | sigma | statistic | p.value | df | logLik | AIC | BIC | deviance | df.residual | nobs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.426 | 0.424 | 14.5 | 155 | 4.62e-51 | 2 | -1.72e+03 | 3.44e+03 | 3.46e+03 | 8.72e+04 | 417 | 420 |